earn how to quantify polymorphic impurities in APIs using X-Ray Powder Diffraction (XRPD) with a step-by-step analytical approach, calibration formula, and a real-world Carbamazepine case study
To quantify polymorphic impurities using X-ray Powder Diffraction (XRPD), a calibration curve must first be established by analysing physical mixtures containing known proportions of the polymorphic forms. This process involves identifying distinctive diffraction peaks for each polymorph, measuring their peak intensities or integrated areas, and plotting these values against the known concentrations. A linear regression equation of the form y = mx + C is then derived, where y represents the measured intensity or area, x is the impurity concentration, m is the slope, and C is the intercept. This calibration curve is subsequently used to determine the concentration of the polymorphic impurity in unknown samples with high accuracy and reproducibility.
Solid-state characterisation is critical to ensure drug efficacy, safety, and stability of an API. One important aspect of this characterisation is the detection and quantification of polymorphic impurities in Active Pharmaceutical Ingredients (APIs). Polymorphs—different crystalline forms of the same compound—can exhibit significant differences in solubility, bioavailability, and stability. Detecting even minor polymorphic impurities is essential to meet regulatory standards and guarantee consistent therapeutic performance.
In this article, I will discuss a stepwise analytical approach using X-Ray Powder Diffraction (XRPD)—a gold-standard technique for characterising crystalline materials—for the quantitative analysis of polymorphic impurities in APIs.
Why XRPD?
X-Ray Powder Diffraction is a non-destructive, highly specific technique that provides direct information about the crystal structure of materials. Unlike other techniques (like DSC or FTIR), XRPD can distinguish between polymorphs based on their unique diffraction patterns, making it ideal for both qualitative identification and quantitative analysis.
Quantification of Polymorphic Impurities in APIs Using XRPD involves the following steps:
Step 1: Sample Preparation
Proper sample preparation is critical. The API must be:
Expert Tip: Poor sample handling can lead to peak broadening, preferred orientation, or inaccurate results.
Step 2: Reference Polymorph Selection
Identify and obtain pure forms of all relevant polymorphs, including:
These references will be used to generate calibration standards.
Step 3: XRPD Data Collection
Set the instrument parameters for optimal resolution:
Ensure that the baseline is flat and peaks are sharp for accurate quantification.
Step 4: Peak Identification
Use software or databases (e.g., ICDD PDF) to match peaks and identify:
Choose non-overlapping peaks for quantification whenever possible.
Step 5: Calibration Curve Preparation
Prepare physical mixtures of the reference polymorphs in known proportions (e.g., 0%, 1%, 5%, 10%, 25%, 50%). Then:
This allows you to establish a quantitative relationship.
Step 6: Sample Quantification
Measure the sample’s XRPD pattern and compare it against the calibration curve to determine:
If advanced deconvolution is needed (e.g., for overlapping peaks), use techniques like:
Step 7: Method Validation
Validate the method according to ICH guidelines:
This ensures regulatory compliance and reproducibility.
Looking to grow your pharma career?
Pick up your course and learn from industry professionals with PharmaGuru’s recognised training programs:
Online Pharma Course (Training)
Objective
To detect and quantify a known polymorphic impurity (Form II) in batches of Carbamazepine API, where Form III is the therapeutically approved and stable form. The study aims to ensure batch consistency and regulatory compliance using X-Ray Powder Diffraction (XRPD).
Background
Carbamazepine, an anticonvulsant and mood-stabilising drug, exists in multiple polymorphic forms (Form I to Form IV). Form III is considered the stable and pharmaceutically acceptable form. However, Form II, a metastable polymorph, can appear during certain crystallisation or milling processes. Even trace amounts of Form II may impact solubility, dissolution rate, and long-term stability.
Materials and Methods
1. Sample and Reference Materials
2. Sample Preparation
3. Instrumentation
4. Calibration Curve Setup
5. Data Analysis
Calibration Curve
| Batch | Peak Intensity at 15.2° 2θ | % Form II (calculated) |
|---|---|---|
| A | 0.00 | < LOD (Limit of detection) |
| B | 0.45 | 1.2% |
| C | 1.00 | 2.8% |
Result Summary
XRPD successfully detected and quantified polymorphic impurities down to 1% concentration, providing a non-destructive and reproducible method for quality control.
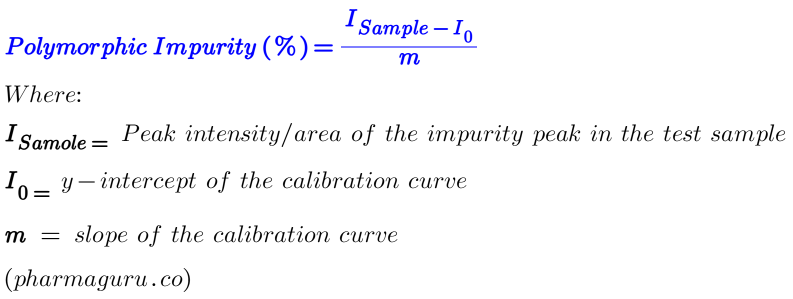
You create a calibration curve by preparing mixtures of known proportions of the impurity with the main polymorph (spiked sample with impurity), such as, 0%, 1%, 5%, 10%, etc., then:
Suppose Slope (m) is 22.5 and Intercept (I₀) is 5.0, then your calibration curve equation is:
I=22.5×C+5.0 (Equation-1)
Where:
If your test sample gives Isample=27.5 then Polymorphic Impurity (C, which is unknown) can easily be calculated from equation 1 in the following way:
27.5=22.5xC+5.0
or C (%)= (27.5-22.5)/5=1%
The quantification of polymorphic impurities in APIs using XRPD is a robust and reliable method when implemented carefully. A stepwise approach—from sample preparation to validation—not only improves accuracy but also ensures regulatory compliance and product consistency.
As pharmaceutical regulations continue to emphasise solid-state control, mastering XRPD-based polymorph quantification becomes a valuable skill for analytical scientists and formulation developers alike.
You may like:
Polymorphs are identified by comparing their unique X-ray powder diffraction (XRPD) patterns, as each polymorph has a distinct crystal structure that produces a characteristic set of diffraction peaks. Additional techniques like Differential Scanning Calorimetry (DSC), Fourier-Transform Infrared Spectroscopy (FTIR), and Solid-State NMR can support identification.
Methods used to determine polymorphism include:
1. X-ray Powder Diffraction (XRPD) – primary method for identifying crystal forms
2. Differential Scanning Calorimetry (DSC) – detects thermal transitions between polymorphs
3. Thermogravimetric Analysis (TGA) – evaluates stability and decomposition
4. Fourier-Transform Infrared Spectroscopy (FTIR) – detects differences in molecular vibrations
5. Solid-State NMR – provides detailed structural information
6. Hot-Stage Microscopy – visualizes phase changes upon heating
These techniques are often used in combination for reliable polymorph identification.
Common techniques include X-ray powder diffraction (XRPD), single-crystal X-ray diffraction, differential scanning calorimetry (DSC), thermal gravimetric analysis (TGA), infrared (IR) spectroscopy, Raman spectroscopy, and solid-state NMR.
Polymorphism affects a drug’s solubility, stability, melting point, bioavailability, and manufacturability. Identifying and controlling polymorphs ensures consistent quality, efficacy, and safety of the drug product.
Polymorphism is tested using XRPD and thermal analysis (DSC/TGA) as primary methods, often supported by spectroscopic techniques (IR, Raman) and microscopy to confirm the crystalline form and detect any polymorphic transitions.
Further reading:
Quick Links